
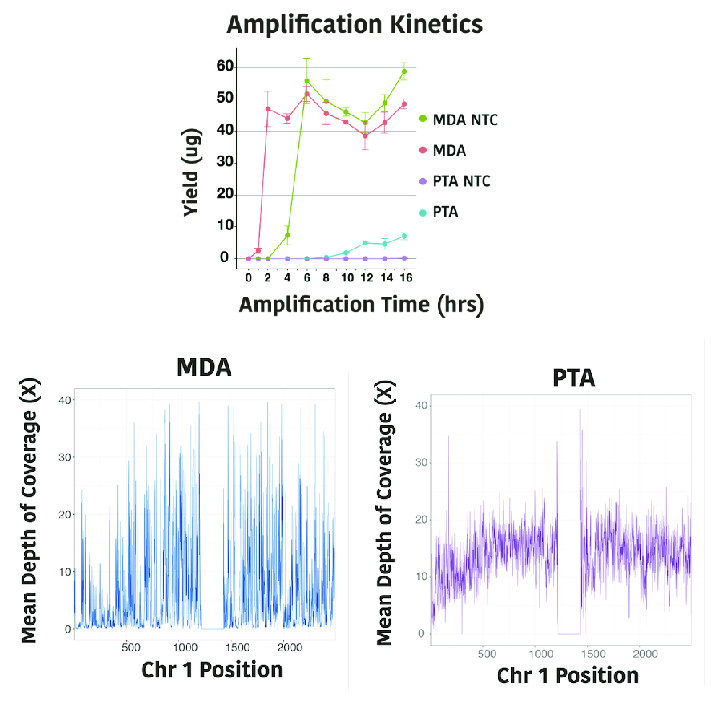
Our lab recently invented a new method, primary template-directed amplification (PTA), for amplifying the genome of a single cell. Our lab and others have established that PTA provides more complete and accurate representations of the original genome than all previously available whole genome amplification methods, enabling us to detect about 90% of small genomic variants with high precision. PTA is being applied to study many different sample types– from low input DNA samples such as ancient DNA to a range of eukaryotic cell studies to more accurately amplifying and assembling the genomes of single microbes. More recently, PTA has been paired with the Smart-seq chemistry to enable accurate genomic variant detection along with transcript quantification in the same cells. We are now working on a method for sparsely marking methylated cytosines to enable genome wide variant calling, transcript abundance, and regional methylation detection from the same cells. In addition to single-cell genomics, our lab is working on wet lab and computational methods to more accurately detect mutations, methylation, and microbial nucleic acid in cell-free DNA samples.
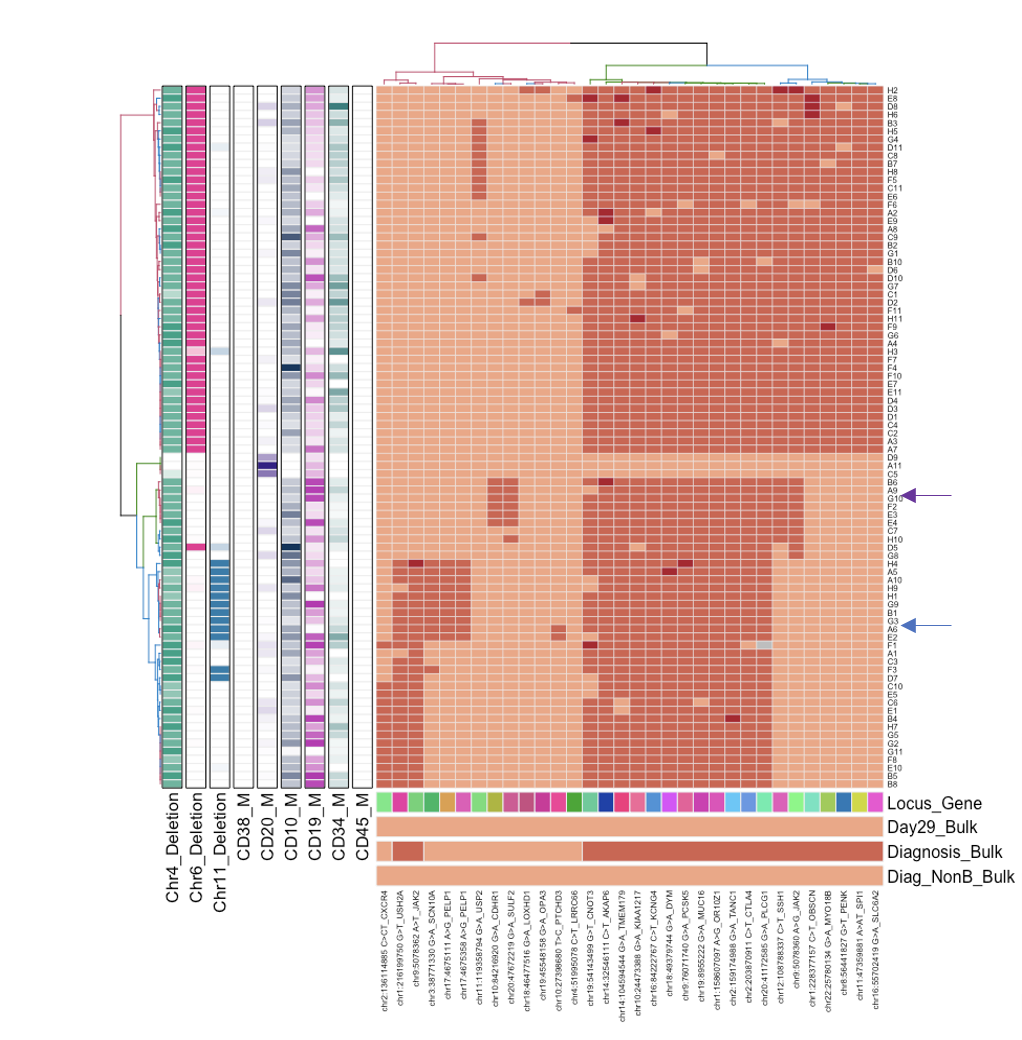
As a practicing pediatric oncologist, I care deeply about understanding why some oncology patients are cured with standard treatment regimens while others have disease recurrence, resulting in poor outcomes. In most patients, it is not that the entire cancer is inherently resistant to treatment, but that a very small fraction of the total population of malignant cells harbor or acquire resistance to many different anti-cancer agents, resulting in a malignancy that is incurable for many patients. Using the single cell methods we and others have developed, we are isolating and sequencing rare persistent cancer cells with the aim of understanding the biological mechanisms of treatment resistance, as well as identifying biomarkers that can serve as diagnostics and therapeutic targets to help guide patients to the most effective and least toxic treatment strategies. We are also complementing these studies with novel methods for identifying mutations, methylation changes, and microbe nucleic acid in cell-free DNA.
All humans develop from a single cell into about 35 trillion cells, acquiring a somatic mutation about once every cell division. In addition, we replace billions of cells daily through ongoing divisions and continue to acquire mutations with exposure to environmental compounds as we age. Although we know this massive intra-tissue cellular genome diversity is present in each of us, little is known about how the mutations in those cells contribute to human diseases. Further, we know that a very small number of cells with somatic mutations evolve into a malignancy, suggesting that specific mutations interact with the epigenetic state of that cell to create disease phenotypes. We are deploying methods developed in our lab to examine the genetic and epigenetic states of the same single cells from normal and diseased human tissues with the aim of understanding the dependent and independent interactions of specific genetic and epigenetic features on abnormal cellular phenotypes. As an example, the video on the left depicts a collaboration with Gerry Grant (Chair of Neurosurgery at Duke) where he implants electrodes into pediatric patients with severe seizure disorder to map out zones for surgical resection. We then remove the tips of the electrodes and are able to sequence almost the entire genome of a single neuronal nucleus in an attempt to understand the underlying genetic changes that result in abnormal brain development.
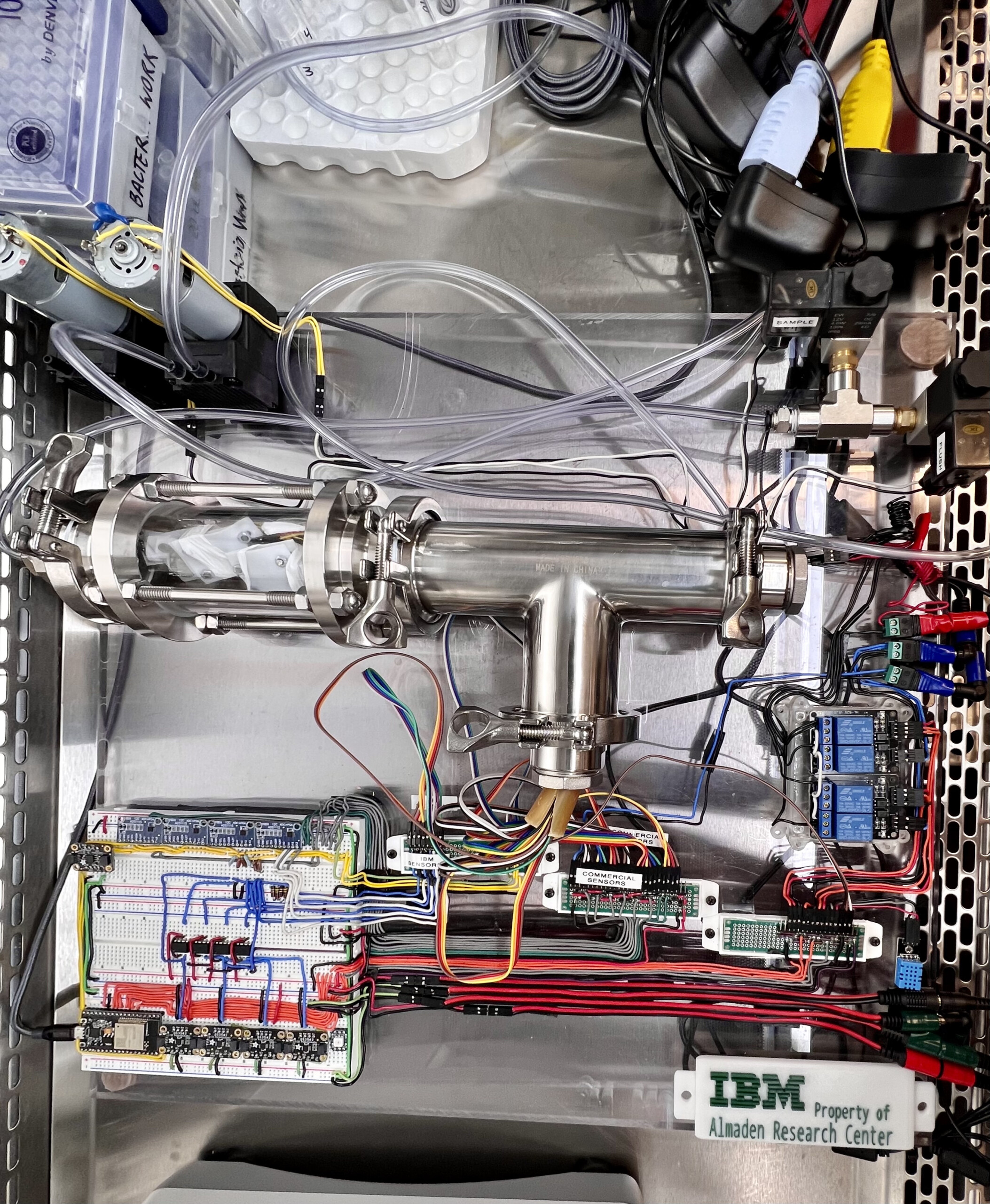
Bacterial sepsis causes about 20% of global deaths annually. In collaboration with scientists and engineers from the IBM Innovation Labs in Almaden, we are developing a novel electronic nose system to detect diagnostic volatile organic compound signatures of bacterial sepsis from human samples. IBM has created a series of novel metal oxide gas sensors with high sensitivity, and have engineered a device to house the sensors, control flow of the incoming samples, and rapidly output a recording of the electrical signals. Although these sensors have the potential to provide extraordinarily high sensitivity, many of the signals are overlapping and nonspecific. Our lab is working to catalogue volatile organic compounds in blood that are specific to patients who have a bacterial bloodstream infection using mass spectrometry with the aim of identifying volatile biomarkers that can be amplified chemically and uniquely interact with the electronic nose system to produce a fast and inexpensive point-of-care test for bacterial sepsis.